INNOVATIONS
Small RNA sequencing and assembly methods revolutionize virus detection
- Nutrition & food security
- Poverty reduction, livelihoods & jobs
Adoption or impact at scale
Taken up by ‘next users’
Start date: 2007 | End date: Ongoing
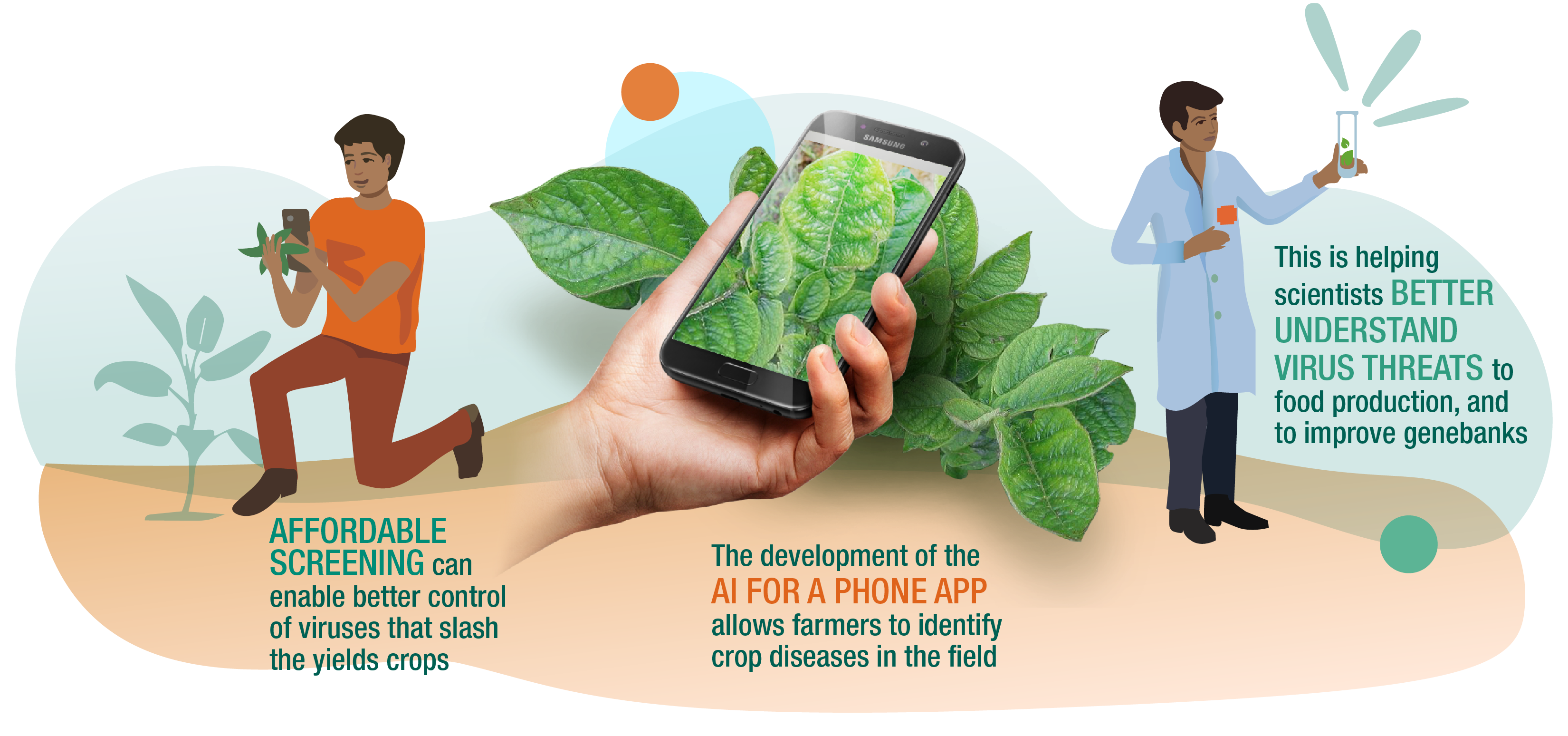
Efficient and accurate detection of viruses in plants and animals is essential for the development of effective strategies to manage disease spread and impact. Conventional detection methods require prior knowledge or sequence information of the potential pathogens, and are not very efficient in detecting novel viruses or virus variants. A new approach, virus discovery by high throughput sequencing and assembly of total small RNAs (sRSA; Kreuze et al., 2009), has proven to be highly efficient in plant and animal virus detection and has been widely adopted by the scientific and disease detection community. This has been enhanced by use of VirusDetect, an automated bioinformatics pipeline operating under Linux developed by Cornell University and CIP in 2017 that can efficiently analyze large-scale small RNA (sRNA) datasets for both known and novel virus identification.

VirusDetect can be applied to detect viruses in various crops, fungi or animals. It has been taken up by researchers in a number of countries, including some African countries , and is being evaluated by CGIAR germplasm health units for adoption as the standard tool to certify virus-free status of clonal crops. Those crops, which include banana cassava, potato and sweetpotato, suffer major yield loss due to viruses because the pathogens are spread via planting material, as well as insect vectors.
The use of sRSA to detect viruses has great potential to enhance efforts to ensure plant health on the level of genebanks, in seed systems for vegetatively propagated crops and in the field.
Bill & Melinda Gates Foundation; European Union (FP7); Howard G. Buffett Foundation; National Science Foundation (USA).
CGIAR Research Program on Roots, Tubers and Bananas; International Institute of Tropical Agriculture; Alliance Bioversity International and CIAT; Boyce Thomson Institute (software); Helsinki University.